
Research


|
Research |

|
Currently, the major research goals in my group focus on answering the following three general questions:
Question 1: What is the correct physical description of a crawling eukaryotic cell and how do biochemistry and biophysics conspire to drive a single cellís motion?
Question 2: How do single cell crawling and cell-cell interactions lead to the complex bulk migration that is observed during wound healing and cancer metastasis?
Question 3: What role do motility and the physical properties of a bacterium play during the transmission and progression of Lyme disease?


|
A complete understanding of the mechanisms by which cells crawl is still lacking, but it is known to entail at least
three separate physical processes: (i) cytoskeletal extension at the front of the cell; (ii) adhesion to the substrate at the cell front and release at the rear;
and (iii) advance of the cell body. In most cells, the cytoskeletal network is composed of actin. The mechanism by which force is generated to drive
translocation of the cell body is still debated. Originally, this force was attributed to an actomyosin system similar to muscle.
However, Myosin II-null Dictyostelium discoideum cells are still capable of translocation, and Myosin IIA-deficient fibroblasts
migrate faster than wildtype cells. Therefore, it is possible that the translocation of the cell during crawling is
driven partially by the dynamics of the actin network without the action of molecular motors. Whatever the role of myosins are in cell motility,
constructing quantitative models for cell motility will require untangling the physics of the cytoskeleton from the action of molecular motors.
My group studies the crawling of sperm cells from the nematode Caenorhabditis elegans. Nematode sperm provide an excellent model system for studying cell crawling in the absence of molecular motors. Unlike most other crawling cells, nematode sperm utilize a cytoskeleton composed of a network of Major Sperm Protein (MSP). MSP forms non-polar filaments, for which no molecular motors have been identified; yet, the motility of these cells still exhibits the fundamental processes of standard crawling motility. In these cells, polymerization at the leading edge is believed to drive the advance of the front of the cell. The molecular level mechanism for adhesion of these cells to the substrate is still unknown. We have proposed a quantitative model that shows that depolymerization of the cytoskeleton may produce the force that pulls up the rear. We use time-lapse microscopy and mathematical models to explore the underlying mechanisms of nematode sperm motility. The figures to the right show DIC images of C. elegans sperm cells and a simulation result that we generated using our mathematical model. The middle figure shows how we extract the boundary of the sperm cells from DIC images. We use an image processing technique developed in our lab to convert the DIC image into a pseudofluorescent image. It is then easy to automatically capture the cell boundary. The movie that is playing on the bottom, shows a simulation of a crawling cell that was done using a simple model for depolymerization-driven motility. We use a moving boundary algorithm that we developed to solve for the shape changes produced by this model. |
|
Lyme disease, which is caused by the spirochete Borrelia burgdorferi, is the most common tick-transmitted illness in the United States.
If untreated, Lyme disease can lead to a wide array of complications typically involving the heart, joints, or nervous system.
It is widely believed that the motility of B. burgdorferi is essential for the pathogenesis of Lyme disease.
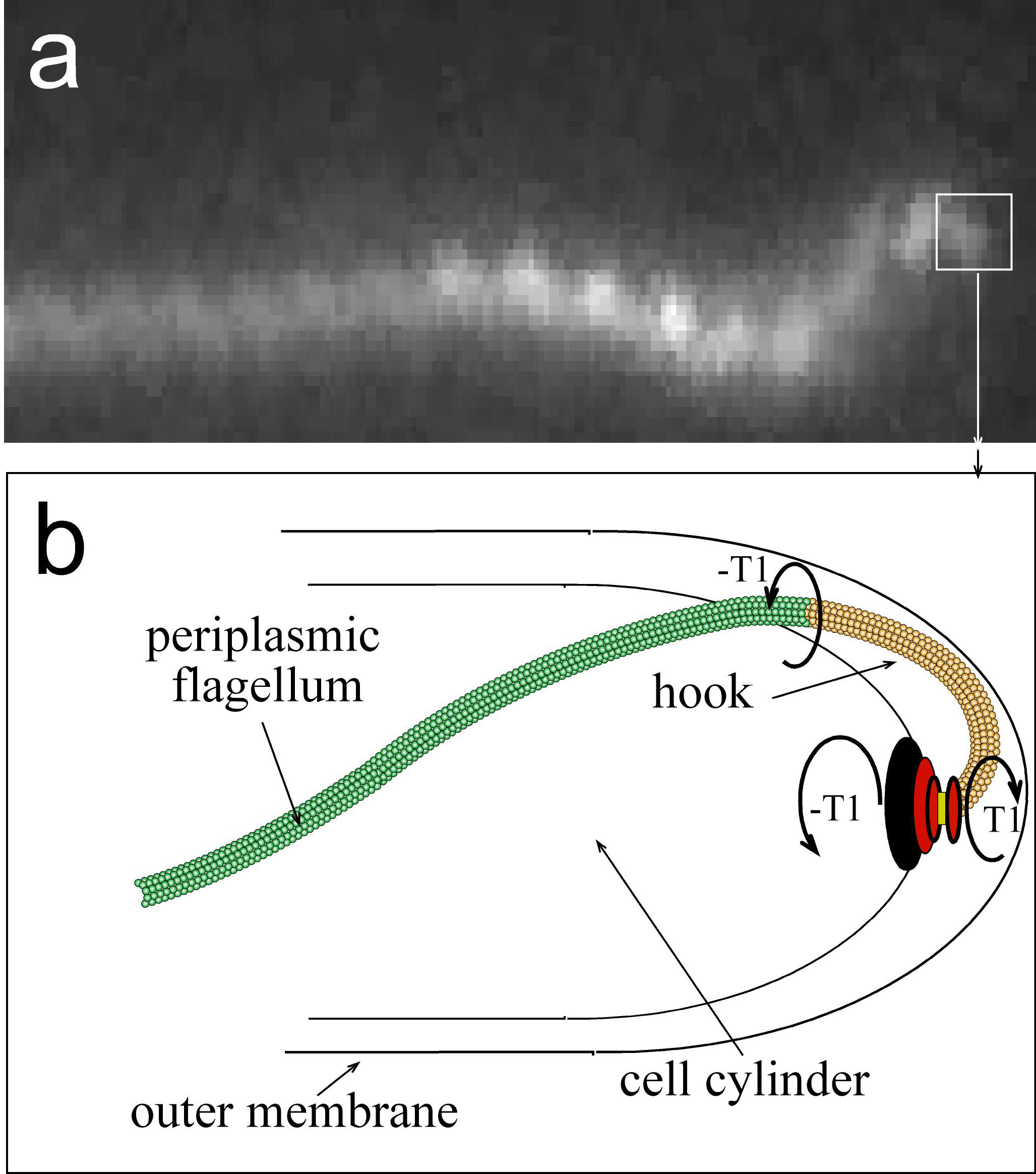
B. burgdorferi swims by rotating helical filaments (flagella) that reside in the periplasmic space
(the space between the outer membrane and the cell wall material). The rotation of these periplasmic flagella against the cell wall leads to
deformations of the cell cylinder, and these deformations exert force against the external environment. The environment pushes back
against the cell, which leads to the net translation of the cell through the environment. Since the cell wall of B. burgdorferi is elastic,
the shape of the cell during swimming is determined by both the force from the flagella and the response force from the environment.
However, since the response force, and therefore the swimming speed, is determined by the shape of the cell, there is a complex interplay
between the environment and the morphology and motility of B. burgdorferi.
The transmission of B. burgdorferi from the tick to its host requires penetration through the tick gut and then into the hemolymph and the salivary gland. Once in the host, B. burgdorferi must navigate through extracellular matrix (ECM) and into blood vessels. To exit the blood vessels, the spirochetes must attach to the vascular endothelium and work their way through tight junctions and back into tissues. Many of these environments are either highly cellular or gel-like, and will respond to applied forces in a different way than would standard media. An added complexity is that B. burgdorferi is able to adhere to cells and ECM proteins, such as collagen and fibronectin. It is therefore likely that during pathogenesis the motility of B. burgdorferi will differ drastically from what is observed in fluid media or in gel-like environments to which the spirochete is unable to adhere; however, all work that has been done to understand the motility of these organisms has been done in fluid or methylcellulose solutions. |

|
| This project is a continuation of a project funded under NIH R01 GM72004, entitled ďAn elastic model for spirochete morphology and motility.Ē In that project, I developed and tested a mathematical model that describes how coupling of the helical, elastic periplasmic flagella to the cell cylinder creates and maintains the shape of some species of spirochetes, such as Borrelia burgdorferi and the Leptospiraceae. The figure to the right shows images of B. burgdorferi and the shape that is computed from our model. Panels (c,g) show that the bacterium is rod-shaped when the helical flagella are not present (d,h). The model also explains the dynamic shape changes that are produced by rotation of the flagella and that drive swimming in the Leptospiraceae. We are currently focusing on B. burgdorferi, as it can be genetically manipulated and its swimming motility and ultrastructure have been well-characterized. We hypothesize that the internal mechanism driving the motility of B. burgdorferi (i.e., flagellar rotation) is largely unchanged when the spirochete moves from one environment to another, but its strategy for motility (i.e., the gross deformations of the cell and how these deformations lead to net translation) is substantially different due to its interactions with the environment. We are, therefore, proposing that the shape; physical parameters, such as the stiffness of the flagella and cell cylinder; and the internal mechanism driving motility in B. burgdorferi, have evolved to allow for directed migration in a wide range of environments in the tick and mammalian host tissues. To this end, our goals are three-fold. First, we want to show that the previously developed model can explain the morphology of cells when cell or flagellar stiffness is modified and that altered stiffness results in decreased motility. Second, we will produce a complete, quantitative understanding of the motility of B. burgdorferi in bulk, gel-like media with an emphasis toward biologically-relevant gels such as collagen networks. Finally, we will use modeling and time-lapse fluorescence microscopy to determine the mechanisms of motility in the tick and in epithelial cell layers. These aims are directed toward moving the current understanding of motility in non-physiological liquid and/or methycellulose solutions to more realistic environments in which spirochetes adhere to cells or ECM in order to complete their enzootic cycle and accomplish their parasitic strategy. In addition, we will determine the physical processes by which these cells are able to penetrate out of the tick and into the host mammal. |
|